Examining complex gill disease in Scottish salmon
These studies sought to build the foundations for improved monitoring, diagnosis, understanding, and management of gill health in Scotland’s farmed salmon. The project has had major scientific, technical, and practical impacts for the salmon sector and research community.
Project team
Partners: Scotland’s Rural College (SRUC), University of Glasgow, Loch Duart, Grieg Seafood Shetland (now Scottish Sea Farms), Bakkafrost Scotland (formerly Scottish Salmon Company), Cooke Aquaculture Scotland, Mowi Scotland, Wester Ross Fisheries (now Mowi Scotland), Nevis Marine, Moredun Research Institute, Salmon Scotland (formerly SSPO)
Wider team: PHARMAQ Analytiq
Impact
This project provided the first standardised and evidence-based definition of complex gill disease, resolving long-standing confusion in terminology and enabling consistent diagnosis. It also delivered the first quantitative estimates of diagnostic test performance under Scottish conditions and produced validated methods for microbiome analysis. For salmon producers, project findings translated into improved management capability.
£2.4m
Total value
Case study
Download PDF
BACKGROUND
In recent years, Scotland’s salmon sector has been affected increasingly by gill health challenges. Across marine farms, inflamed and damaged gills have been observed which can lead to respiratory issues, reduced growth, emergency harvests, and significant economic losses.
The cause of gill disorders in marine-farmed salmon is complicated and, often, gill disease is believed to be the result of many interacting factors. These factors include infectious agents such as amoebae, bacteria and viruses; and environmental stressors such as harmful plankton, in adverse water temperature, or poor water quality. When no single primary causative agent can be identified, the condition is known as complex gill disease By 2017, the term was being used widely but without a clear or consistent definition, making it difficult to compare data or design effective management strategies.
Recognising the scale and complexity of the problem, the Sustainable Aquaculture Innovation Centre (SAIC) announced its funding of the ‘Gill Health in Scottish Farmed Salmon’ project, which brought together academic researchers, diagnostic experts, and industry partners from across Scotland. It aimed to produce an evidence-based understanding of gill disease from two complementary strands of work:
- Work Package 1: investigation of the epidemiology of CGD, identified risk factors, diagnostic performance, and prevalence.
- Work Package 2: characterisation of the community of microorganisms living on fish gills, or the microbiome, to understand its relationship with disease.
Together, these studies sought to build the foundations for improved monitoring, diagnosis, understanding, and management of gill health in Scotland’s farmed salmon.
AIMS
The project’s overall goal was to understand what complex gill disease is, how to recognise it, what causes it, and how to control it. Specifically, it aimed to:
- Define CGD consistently so that farmers, veterinarians, and researchers can identify it in the same way and compare data and putative causes related to CGD outbreaks;
- Measure the prevalence of CGD in the Scottish fish farming sector and how its prevalence has changed over time;
- Assess the accuracy of existing diagnostic tests and possible improvements;
- Identify biological, environmental, and management factors that increase the risk of CGD;
- Explore new biomarkers and microbiome indicators that could help detect or predict CGD;
- Quantify the effect of CGD on fish welfare and production.
These objectives were designed to improve understanding and management of gill health by supplementing experience-based observation with data-driven management supported by strong scientific evidence.
PROJECT METHODS AND OVERVIEW
Defining complex gill disease
One of the project’s first tasks was to establish a clear, standard definition of CGD, as there was no consistent understanding of what the term meant. The research team reviewed international literature, surveyed Scottish and Irish fish health professionals, and held a workshop with leading European fish histopathologists. They also drew on new field data to test and refine the definition.
The review and expert consultation recognised seven main categories of marine gill disease: amoebic, parasitic, viral, bacterial, zooplankton-related, harmful algae-related, and chemical/toxin-related. When one or several of these occurred together with unspecific pathology, the condition was complex gill disease (CGD).
Prospective longitudinal study
From September 2018 to June 2020, researchers conducted a prospective longitudinal study including intensive sampling on eight marine production sites and their pre-transfer freshwater locations, managed by six salmon companies. Two pens per site were sampled approximately every two weeks, from seawater transfer to harvest. At each visit, eight fish per pen were examined: half using non-lethal gill swabs and visual assessments, and half by adding terminal sampling for detailed tissue and blood sampling. All procedures were done by trained farm staff or fish health specialists under approved animal welfare protocols.
This prospective dataset provided a unique consistent, high-quality record of gill condition, environmental factors, and potential pathogens over time, capturing 18–34 sampling points per site and creating a detailed longitudinal picture of disease progression and associated variables that is still in use today in various follow-up (inter-)national projects. This fieldwork experienced some interruptions from early 2020 due to COVID-19.
Retrospective dataset
To complement the above work, a retrospective dataset was assembled from industry partners, containing from 2013 to 2019. This included production and health data such as mortality, biomass, current speeds, seabed type, net-cleaning and treatment dates, test results for amoebic and proliferative gill disease, and cleaner fish information. While these data were less standardised compared to the 2018-2020 dataset, they covered hundreds of site-cycles across Scotland, offering a comprehensive picture of national gill health trends.
Using the CGD definition, the team analysed the new and historical data to estimate disease prevalence, assess diagnostic accuracy, and model how many fish need to be sampled for reliable site-level results. Statistical approaches accounted for and quantified clustering effects, because samples from fish within the same pen tend to be more similar to each other than samples from fish from different , and understanding the degree of similarity can inform surveillance strategies (e.g. how many fish to sample from how many pens).
To interpret test results correctly, it is essential to know how well each test correctly identifies presence or absence of infection and disease. Using data from the prospective study and Bayesian statistical models, the researchers estimated diagnostic sensitivity (how well a test detects true positives) and specificity (how well it detects true negatives) for the most common tests: gross scoring, qPCR, and histopathology.
Biomarker and microbiome investigations
Alongside epidemiological work, the project examined potential biomarkers of disease using blood chemistry, histology, and molecular detection of suspected pathogens such as Desmozoon lepeophtherii, salmon gill poxvirus (SGPV), and Candidatus Branchiomonas cysticola.
Work Package 2 focused specifically on the gill microbiome. Researchers developed and validated DNA extraction and sequencing methods to identify bacteria on the gills of both healthy and diseased fish. They examined how environments influenced the microbiome before seawater transfer and how it thereafter changed . Samples were collected as part of the prospective study described earlier. This was the first coordinated effort to describe microbial communities on salmon gills across multiple commercial farms in Scotland.
RESULTS
Standardising definitions and testing for CGD
The project produced several major advances in the understanding complex gill disease.
The first was a practical, data-driven case definition for CGD. By combining histopathology, gross gill scoring, and molecular testing, the team created a framework that could be applied consistently across farms and studies.
CGD was ultimately defined as: a gill disorder characterised by non-specific pathological changes, such as epithelial thickening, lamellar fusion, inflammation, necrosis, and haemorrhage, without an obvious identifiable cause.
The project introduced three diagnostic “stringency levels” (mild, moderate, severe), based on combinations of gross gill scoring and histopathological results. A fish was classed as CGD-positive if it tested positive on at least one of three standard diagnostic measures: histopathology, gross gill scoring, and molecular testing.
For AGD, PCR testing proved highly sensitive and specific, while histopathology was less sensitive but very reliable for confirming disease. For CGD, gross total-gill scoring had the highest sensitivity (around 0.9), while histopathology again offered strong specificity but lower sensitivity. These findings confirmed that not all tests give the correct result at all times and that using several tests in combination gives the most accurate diagnosis. The project’s insights enables more reliable assessment of true prevalence from routine monitoring results, a better understanding of what a test result means, and which test to use in which circumstance.
Prevalence and trends
Using this definition, the researchers provided the first quantitative estimates of CGD prevalence in Scottish aquaculture. In the prospective dataset (2018-2020), 29% of pens showed mild CGD, compared with 15% in the retrospective data (2013-2019), which spanned earlier years., around 2% and 0.2% respectively in both studies (see Table 1).
- Overall prevalence of CGD based on the pen average of PGD score
|
Stringency |
Case definition based on a pen average of the PGD score |
Prospective study (2018-2020) |
Retrospective study (2013-2019) |
|
Less |
≥ 1 |
29% |
15% |
|
Medium |
≥ 2 |
2% |
2% |
|
Severe |
≥ 3 |
0.2% |
0.2% |
|
N (pens) |
N |
473 |
121,716 |
Over time, patterns shifted: amoebic gill disease declined from 2013 to 2018, but CGD increased over the same time period, especially in 2017–2018. Gill ds tended to peak between late spring and autumn, though timing varied across the eight sites in the prospective study (see Figure 1). Possible reasons considered were: AGD treatments had become more effective, multifactorial CGD had emerged as the dominant gill health challenge, and .
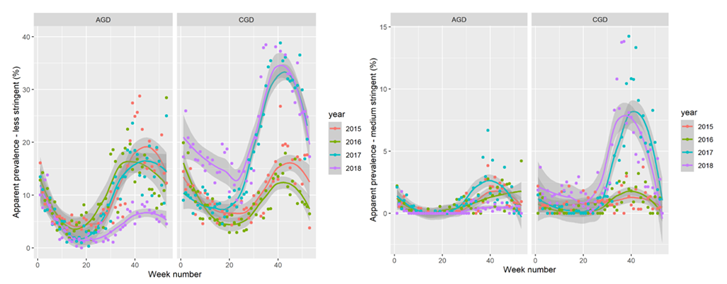
Figure 1. Prevalence of less and medium stringent AGD and CGD throughout a year, based on retrospective data
To support sampling, the team developed an online sample-size calculator, freely available from SRUC, to help farmers and vets decide how many pens and fish to sample for reliable monitoring, depending on expected disease levels and desired accuracy. This tool now supports more efficient and standardised gill-health surveillance across the sector.
Biomarker analysis identified several indicators associated with CGD. Physical signs such as bleeding gills, operculum deformities, scale loss, and visible blood during anaesthesia were strongly linked to . Molecular detection of D. lepeophtherii, SGPV, and N. perurans was also associated with higher disease risk. In histopathology, cellular necrosis and inflammation . Blood analysis suggested that higher lymphocyte counts corresponded to increased risk, while higher heterophil levels were protective. were linked with CGD, while higher urea appeared to indicate better health. These biomarkers represent valuable leads for future development of diagnostic or prognostic tools.
Risk-factor analyses confirmed that environmental and management conditions play key roles in CGD. Harmful plankton blooms, both phytoplankton and zooplankton, were often with spikes in gill irritation and disease. Other factors such as water temperature, current speed, and net-cleaning protocols also influenced outcomes. Sites experiencing repeated CGD outbreaks showed higher mortality, slower growth, and poorer feed conversion, illustrating the direct production impacts of poor gill health.
Microbiome dynamics
Work Package 2 revealed that the gill microbiome is highly dynamic. Hatchery system is a strong driver of the gill microbiome, with the gill microbiome of fish influenced by whether they were reared in flow-through, loch or recirculating systems.
Once in the marine environment, the microbiomes became more similar, but continued to change over time, with a strong cyclical seasonal trend. After a period in the marine environment a sharp dysbiosis of the microbiome occurred with a decrease in species richness, the number of different bacteria on the gills, followed by a recovery. This was not related to the outbreak of disease, but was driven by the dominance of a single organism, Ca Branchiomonas cisticola. It was found in all fish across all sites and times.
The gill microbiomes of gill diseased fish and non-diseased fish were compared before-during and after outbreaks of disease. While no obvious signature of gill disease was observed in the microbiome, CGD was a statistically significant variable; and diseased fish had a smaller core microbiome.
IMPACT
This project had major scientific, technical, and practical impacts for the salmon sector and research community.
Scientifically, it provided the first standardised and evidence-based definition of complex gill disease, resolving long-standing confusion in terminology and enabling consistent diagnosis. It also delivered the first quantitative estimates of diagnostic test performance under Scottish conditions and produced validated methods for microbiome analysis. The new online decision-support tools, including the sample-size calculator and diagnostic interpretation guide, are already used by fish-health teams and researchers to plan and evaluate surveillance programmes.
For salmon producers, project findings translated into improved management capability. Farmers can now sample fish for gill disease being better informed, interpret test results more accurately, and better understand the role of environmental conditions in disease outbreaks. Recognising that often results from multiple interacting factors reinforces the need for integrated health management, combining water-quality monitoring, plankton observation, stock handling, and site planning.
The microbiome research laid the groundwork for potential new approaches to disease prevention, such as probiotic strategies or microbiome-based health indicators, that could support long-term resilience in salmon farming.
The project also demonstrated the value of developing trusting and collaborative relationships between and within academia and industry. Nearly all major Scottish salmon producers contributed data, allowing for national-scale analysis and a shared evidence base, to help solve a shared challenge.
The findings of this research will continue to guide innovation, improve fish welfare, and support the sustainable growth of one of Scotland’s most important food sectors.
Additional information
- Epidemiology of marine gill diseases in Atlantic salmon (Salmo salar) aquaculture: a review - Boerlage - 2020 - Reviews in Aquaculture - Wiley Online Library
- SRUC sample size calculator
- Field evaluation of diagnostic sensitivity (DSe) and specificity (DSp) of common tests for amoebic gill disease (AGD) and complex gill disease (CGD) in cultured Atlantic salmon (Salmo salar) in Scotland using Bayesian latent class models - ScienceDirect
- Hatchery type influences the gill microbiome of Atlantic farmed salmon (Salmo salar) after transfer to sea | Animal Microbiome
- The role of the microbiome in the gill health of farmed Scottish Atlantic salmon - Enlighten Theses