Use of serum biomarkers for early differential diagnostics of cardiomyopathies of Atlantic salmon: field and challenge assessment
The project expanded the knowledge for cardiomyopathies’ dynamics and physiological impacts, as well as further refining the SPARCL™ assays and reaching a commercial level. Although biomarkers are not stand-alone diagnostic or prognostic tools, they can enrich the assessment of a farmed fish population.
Project summary
Partners: University of Glasgow, Cooke Aquaculture Scotland, Life Diagnostics Ltd, Benchmark Ltd, Moredun Research Institute, University of Edinburgh.
Project leads: Prof. David Eckersall, Dr Francesca Riva, Philippe Sourd, Andrei Bordeianu, Dr Christopher C Chadwick, Hooman Moghadam, Dr Kim Thompson, Dr Jorge Del-Pozo.
Impact
£408,838
Total value
Case study
Download
BACKGROUND
Cardiac viral diseases are a major cause of economic losses in Atlantic salmon aquaculture in Scotland and Norway. These include pancreas disease (PD) caused by salmonid alphavirus (SAV), cardiomyopathy syndrome (CMS) caused by piscine myocarditis virus (PMCV), and heart and skeletal muscle inflammation (HSMI) caused by piscine orthoreovirus (PRV).
These diseases can occur concurrently and lead to lesions of the myocardium and/or skeletal muscle. In Norway, CMS, PD, and HSMI have been identified as the three most important viral diseases, and CMS and HSMI are the second and third cause of economic losses after sea lice. Currently, diagnosis of these diseases is based on clinical signs, histopathology and RT-qPCR, which require lethal sampling.
In the previously SAIC-funded CMS project, cardiac troponin C (cTnC), a biomarker for cardiac and slow-twitch skeletal muscle damage, and skeletal troponin C (skTnC), a biomarker for fast-twitch skeletal muscle injury, were identified and evaluated as indicators of CMS infection. Specific antibodies were produced against the markers and incorporated into two assay platforms; ELISA and SPARCL. SPARCL assays were run on a portable VetBio-1 luminometer. Using these assays, it was possible to confirm significantly higher serum levels of cTnC in CMS diseased salmon, while levels of cTnC and skTnC both increased in PD positive fish (ten-fold higher than was noted in CMS positive fish).
ORIGINAL AIM
This project aimed to provide the sector with a serum biomarker panel to enable practical and early differential diagnostics of CMS, HSMI and PD by validating the use of cTnC and skTnC SPARCL™ assays of early/subclinical cardiomyopathies in the field, evaluating longitudinal changes in cTnC and skTnC serum levels in controlled challenge, and developing additional SPARCL™ assays for novel candidate biomarkers.
WORK DONE
Validation of the use of cTnC and skTnC SPARCL™ assays for differential diagnosis of early/subclinical cardiomyopathies in the field
To validate the use of cTnC and skTnC SPARCL™ assays, a six-month study monitored salmon health across four farms. The project investigated these biomarkers aiming to assess the diagnostic and prognostic values, already linked to PMCV and SAV diseases on the previous SAIC-funded project.
Sampling started three months after seawater transfer from four Cooke Aquaculture Scotland farms, chosen for their susceptibility to PMCV and SAV. Samples of sera were sent to Moredun Research Institute (MRI) as aliquots and distributed to University of Glasgow (UoG) and Life Diagnostics Ltd Inc. (LD) for biomarker assay, PatoGen for RT-qPCR, and AFBI for serology. Additionally, sera from healthy salmon were collected for comparison.
For the longitudinal sample set, 756 serum samples were collected over six months, with 24 randomly chosen salmon from each site every month. During sampling, fish were caught and anaesthetised, approximately 5ml of blood collected, and serum aliquots immediately frozen for transport and subsequent analysis.
An outbreak sample set, comprising terminal samples including serum, was collected from fish showing clinical symptoms of CMS and from randomly selected healthy fish. Cardiac tissue samples were collected and fixed for RT-qPCR and histology analyses.
A subset of serum samples was chosen for proteomic studies comprising nine randomly selected serum samples from each of the clinically healthy CMS and PD outbreak groups for quantitative tandem-mass-tags (TMT) proteomics.
Additionally, cardiac, and skeletal muscle samples from the previous SAIC-funded project were used to develop and assess an immunohistochemistry (IHC) assay using a rabbit polyclonal antibody to PMCV. This subset included samples from CMS diseased salmon and clinically healthy salmon, previously characterised by RT-qPCR.
All samples from the longitudinal study were analysed using the SPARCL™ immunoassay, developed by Life Diagnostics in the previous SAIC project, as well as by ELISA, to measure cTnC and skTnC content.
Viral cardiomyopathy outbreaks, identified by clinical signs, occurred during the sample collection, including PD and CMS. PD outbreaks showed substantial cTnC increases, while CMS outbreaks had lower cTnC concentrations than in PD outbreaks but were elevated compared to healthy salmon. In PD-affected pens, cTnC concentrations did not return to pre-infection levels during the study, with persisting elevation in subsequent CMS outbreaks.
Serum concentrations of skTnC exhibited large increases during PD outbreaks, mirroring the patterns observed for cTnC. SkTnC concentrations were 2-10 times higher than cTnC, as seen below, likely attributed to the larger tissue mass of fast-twitch skeletal muscle compared to cardiac and slow-twitch skeletal muscle. In the CMS outbreak there was no difference in skTnC concentration from that found in healthy salmon.
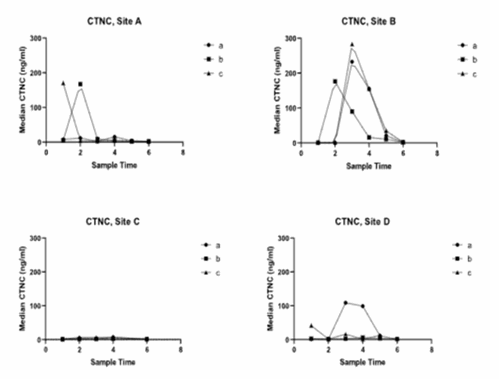
 cTnC values
cTnC values
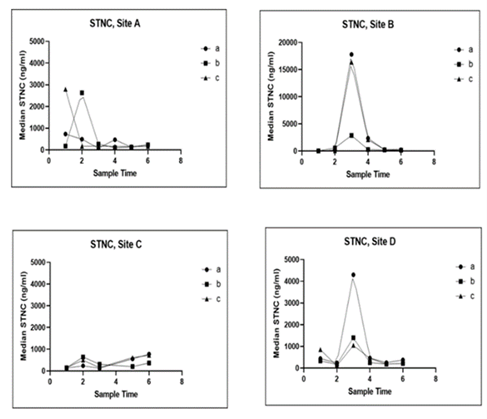
SkTnC values
PD leads to substantial increases in cTnC and skTnC in serum, while CMS causes a comparatively low but significant increase in the former. This suggests cTnC and skTnC are valuable biomarkers for PD. However, distinguishing CMS outbreaks from sites in PD recovery may pose a challenge. Combining these biomarkers with other health assessment tools and exploring secondary biomarkers may enhance differentiation between CMS and PD.
As mentioned before, chosen serum samples from the longitudinal study were sent to PatoGen for RT-qPCR analysis, covering PRV, SAV, and PMCV detection. Serum samples were also sent to AFBI for serological analysis on SAV. based on neutralising antibodies (NT), indicating previous exposure to SAV.
Three of the moribund fish sampled at one of the sites showed positive PMCV levels in sera and, although sera from healthy fish were not tested, testing heart samples revealed that all healthy and moribund fish at this site were positive for PMCV. Additionally, all healthy and moribund fish tested positive for PRV, with one moribund fish also positive for SAV. Fish from an additional site were all negative for all three viruses in both sera and heart tissue.
Fish from some sites displayed detectable levels of NT antibodies, indicating exposure to SAV. Despite exposure, most fish in these sites did not have active viremias. Positive viremias were observed only in two of the three sites, corresponding to known SAV outbreaks. Peak and low cTnC levels showed clear differences between pens within the same site exposed to the same level of viremia, while in another site, peak cTnC and skTnC levels were observed one month after positive viremias.
Heart and muscle histology samples were collected from one of the sites during a clinically detectable outbreak to provide additional insights into diagnosing these specific diseases. Samples consistent with a diagnosis of CMS underwent cardiac tissue scoring based on a scoring system ranging from 0 (no lesions) to 3 (severe lesions).
All samples taken during the farm outbreak of CMS, both clinically diseased and clinically healthy fish, exhibited histological lesions consistent with CMS, with scores ranging from 1-2 for clinically healthy salmon and to 2-3 for clinically diseased salmon. Multifocal hepatocellular necrosis was observed in 6 of 10 salmon with clinical CMS, absent in clinically healthy fish. Mild chronic focal myositis was noted in both groups. Additionally, RT-qPCR assays revealed detectable PCMV in all samples, most having PRV at higher Ct values. One diseased sample showed detectable SAV by RT-qPCR.

Histology results from outbreaks compared to cTnC and RT-qPCR
These results align with clinical interpretation and higher cTnC serum concentrations in clinically diseased salmon. Interestingly, mild myositis was observed in both groups, indicating underlying PRV infection, confirmed by a positive RT-qPCR in most fish. A single fish even showed detectable SAV RNA. No significant correlation was found between PCMV Ct values and cTnC serum concentrations. Higher cTnC levels were observed only in fish with the highest histological scores, suggesting that cTnC serum levels are inadequate for predicting viral load and poorly predict cardiac lesion severity at lower score levels. These results underscore the complexity of field outbreaks.
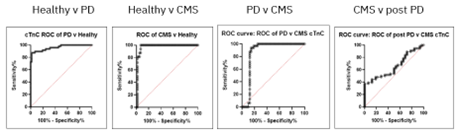
Sensitivity and specificity analysis, as seen below, indicates that cTnC can differentiate PD vs. healthy; CMS vs. healthy; and PD vs. CMS, but when CMS samples were compared to samples from sites with prior outbreaks of PD, but after returning to clinically healthy status (post-PD) marginal differentiation was found.

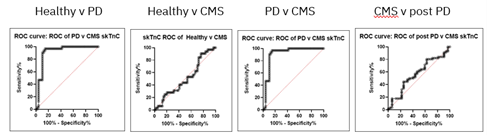
Likewise, sensitivity and specificity analysis, as seen below, suggest skTnC can differentiate PD vs. healthy; and PD vs. CMS; but not CMS vs. healthy or CMS vs. post-PD in aspects not associated with muscle myofibril proteins.

Evaluation of longitudinal changes in cTnC and skTnC serum levels in controlled challenge with viruses causing cardiomyopathies
In parallel to the field study, a controlled challenge test was carried out by Benchmark Genetics in VESO Vikan, Norway, aimed at elucidating cardiomyopathies’ pathogenesis. These tests exposed healthy fish to the virus, monitoring disease progression, impact on fish health and identifying resistant individuals. Close monitoring for clinical signs and longitudinal sampling provided insights into viral load, histopathological heart changes and blood biomarker modifications. While the original plan included challenges for SAV and PRV, samples were only obtained for PCMV.
After acclimatisation, all fish were anesthetised and intraperitoneally inoculated with PMCV and a control group kept separate. The challenge spanned nine weeks, with heart tissue, blood serum and formalin-fixed samples collected at 0, 2, 5, 7 and 9 weeks post-infection (wpi).
The analysis of serum samples by SPARCL™ assay was to determine the effect of the infection on cTnC concentration on serum. Significant differences in cTnC serum concentrations were observed over time. Specifically, there was a sequential increase, significant in post-hoc comparisons between baseline at 0 wpi and 7 and 9 wpi. The mean cTnC concentration increased from 0.97 ± 0.69 ng/ml before infection to 5.7 ± 1.2 ng/ml in the nine-week challenge, as seen below.

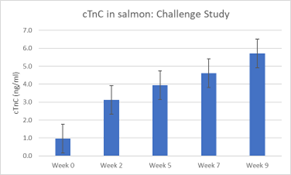
This moderate rise in cTnC was supported by histopathology scoring. Although the increase was limited compared to outbreak cases from the longitudinal study, it indicated damage to cardiac or slow-twitch skeletal muscle due to PMCV infection.
The rationale for the histopathology analysis of challenge samples was to determine the effects on heart, muscle, and other tissues. Lesions were first detected at week five, with higher severity scores increasing over time. Significant differences in cTnC concentrations were found between samples with no lesions (score 0) and those with lesions (scores 1 and 2), but no significant differences between scores 1 and 2.
Overall, these results are promising, indicating that cTnC may moderately predict the presence of histological lesions in a controlled challenge. The observed temporal pattern and significant differences in cTnC serum concentrations between salmon with and without CMS lesions support this finding. A larger sample size and longer challenge may be needed to detect significant differences between histological scores.
Developing additional SPARCL™ assays for novel candidate biomarkers
For the development of four additional SPARCL™ assays for novel candidate biomarkers, antibodies were developed based on peptide sequences specific to chosen biomarkers in salmon or Rainbow trout. ELISA and SPARCL™ assays were then developed for these potential biomarkers. After ensuring the precision and accuracy of the immunoassays through initial validation, samples from the longitudinal studies were assayed for biomarker concentrations. In general, the assays showed low levels for CMS and PD, with no significant differences between samples from healthy or CMS affected fish.
Antibodies were raised and used to develop immunoassays for various biomarkers, including haptoglobin, growth/differentiation factor 15 (GDF-15), cathelicidin, and Complement C1q-LP3. However, assays for salmon retinol binding protein and leptin were not viable for disease diagnosis, being unable to differentiate healthy samples from any of the samples from fish with infectious diseases under study.
Novel biomarkers developed in the project were evaluated in trout by collaborating with the National Centre for Cool and Cold Water Aquaculture (NCCWA), US. Haptoglobin, cathelicidin and complement C1q-LP3 showed increased levels during bacterial infection of trout, with greater responses in susceptible Rainbow trout. Further research is needed for broader application and studying immune response pathways in salmon and trout.
Identification of putative biomarkers of CMS and PD with a quantitative proteomic platform
This section is an additional value to the project, not included in the original proposal. As the issue of differential diagnosis between CMS and PD arose, a quantitative investigation was proposed using samples from the longitudinal study. This involved comparing samples from natural outbreaks of these infections and samples from a disease-free site. The goal was to identify serum protein biomarkers specific to CMS, distinct from PD, or exhibiting a pattern indicative of CMS rather than PD recovery.
Proteomic analysis employed a gel-free tandem mass tag (TMT) quantitative proteomics approach to assess the impact of CMS, PD, and healthy serum samples. Selected differentially expressed proteins identified by the proteomics were further investigated through Western Blot. Primary antibodies were incubated with blots overnight, washed in TTBS and incubated with relevant secondary antibodies. ECL Reagent was applied, blots exposed on X-ray film, and antibody abundances quantified.
After proteomics and initial deconvolution and analysis, a number of proteins exhibited significant increases or decreases when comparing the CMS group to the healthy group. Notably, proteins such as Ig kappa chain V-III region VG precursor and gamma fibrinogen showed significant increases in the CMS group, while apolipoproteins exhibited decreases.

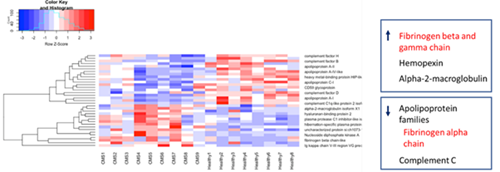
Heat map of proteins showing maximum change in comparison of CMS and healthy samples
In the comparison of the PD group to the healthy group, notable increases in abundance of glycolytic enzymes such as triose phosphate isomerase B and pyruvate kinase, along with decreases in apolipoproteins and albumin, were observed. Principal component analysis of the proteomic analysis results showed a distinct cluster for PD samples, and while CMS and healthy groups were separate, there was some overlap. The Ig kappa chain V-III region VG precursor showed a pattern with greater increase in CMS than healthy and PD samples but antibody for this protein was not available for further analysis.

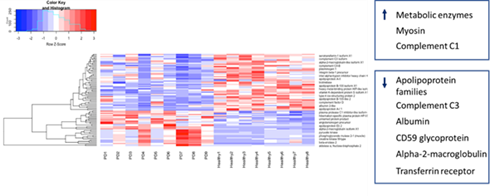
Heat map of PD versus healthy
Antibodies to human gamma FB cross-reacted in a western blot with samples from CMS, PD, and healthy groups. Although bands appeared at the expected 52 kDa, additional higher and lower bands were observed. Further research is needed to explain the patterns, but a biomarker panel including gamma FB and other biomarkers may hold diagnostic and monitoring value for CMS and PD in salmon.
It is possible that the potential biomarkers identified in the proteomics study could aid in the differential diagnosis of PD and CMS, especially if concentrations return to healthy levels after a PD outbreak and is increased in subsequent CMS.
Recently published follow-up research (Riva et al., 2025) confirmed and extended these proteomic findings. Using nine serum samples per group from CMS, PD and healthy salmon, a total of 674 proteins were identified, with 57 classified as differentially abundant. Validation assays demonstrated that β- and γ-fibrinogen were significantly elevated in CMS and PD compared to healthy samples. Importantly, calculation of the fibrinogen-to-skTnC ratio provided clear discrimination between CMS and PD, due to the very high skTnC levels in PD fish compared to CMS. Western blot analysis supported the proteomic results, showing distinct fibrinogen bands in CMS serum compared to healthy fish.

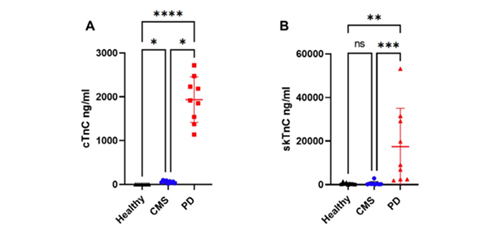
Concentrations of cTnC (A) and skTnC (B) in serum from healthy Atlantic salmon and those with CMS or PD
In PD, the proteomic profile showed a stronger systemic response, with high abundance of glycolytic enzymes (including creatine kinase and enolase), alongside reduced apolipoproteins and albumin. In CMS, changes were more subtle but included increased fibrinogen, Ig kappa chain proteins, and specific alterations in coagulation- and immune-related proteins. PCA confirmed PD formed a distinct cluster, while CMS overlapped with healthy but with some outlier samples separable.
Overall, the proteomic analyses from this project, together with the validation study (Riva et al., 2025), indicate that fibrinogen, particularly when combined with troponins, is a promising biomarker for CMS, enhancing the potential for differential diagnosis from PD.
Development of an IHC assay for PCMV detection in tissue sections
Another additional piece of valuable research was the development of an IHC assay for PCMV detection in tissue sections using antibodies, raised by Life Diagnostics, against PCMV. This IHC assay will be tested further for specificity/sensitivity vs a CMS case definition including histological diagnosis and RT-qPCR, as well as for correlation with these two parameters. Preliminary results suggest that this assay would be suitable for the diagnosis of PCMV infection in samples where the histological presentation may be subtle or unclear and able to identify the presence of the virus in tissue, correlating well with PCMV yes/no in RT-qPCR. However, it may not correlate consistently with the severity of CMS associated lesions or with PCMV load in tissue. This further testing will be part of a student project at the University of Edinburgh.
CONCLUSIONS
The project has successfully established cTnC and skTnC as diagnostic biomarkers for PD and CMS in salmon, providing valuable insights into disease concentrations. These biomarkers, particularly in monitoring disease progression, may aid in monitoring PD recovery and offer potential for differential diagnosis. The findings address ongoing challenges in PD and CMS, contributing to industry advancements, reducing costs, and improving animal welfare.
The analysis of cTnC and skTnC concentrations in salmon serum from the longitudinal study revealed that healthy salmon show lower cTnC concentration compared to skTnC.
SAV infection significantly increases both cTnC and skTnC concentrations, making both biomarkers valid for SAV infection and PD.
PMCV infection and CMS leads to higher cTnC concentration than in healthy salmon but does not increase skTnC concentration, making it an unsuitable biomarker for CMS, lacking differentiation between healthy salmon and with CMS.
Post-PD, cTnC concentrations remain elevated for up to six months, posing a challenge in differentiating PMCV infection, making cTnC a marginal biomarker for PMCV infection and CMS, struggling to fully differentiate CMS from a previous PD. However, monitoring recovery progress could be valuable. Post-PD, skTnC concentrations return to pre-infection levels in 2-3 months.
The developed serological methods offer non-lethal, cost-effective early diagnosis of CMS/PD/HSMI. The SPARCL™ system’s quantitative results provide rapid diagnostic information, enhancing prevention and mitigation strategies. The immediate deployment of SPARCL™ assays in the field is currently feasible and can be used as an on-farm tool to complement existing diagnostic tools. For companies implementing a monitoring programme, the addition of specific biomarkers allows a clearer health assessment of stocks, reducing cardiomyopathy-related losses.
While most additional biomarkers investigated did not provide further diagnostic information beyond cTnC or skTnC, proteomic analyses identified fibrinogen as a valuable candidate. Validation in follow-up research (Riva et al., 2025) confirmed that fibrinogen, particularly when used in combination with troponins, enhances differential diagnosis between CMS and PD. This represents a meaningful step forward in developing a robust biomarker panel for cardiomyopathies in Atlantic salmon.
Publications
Serum Biomarkers in Atlantic Salmon for Differential Diagnosis of Cardiomyopathy Syndrome and Pancreas Disease: Proteomic Identification of Serum Fibrinogen to Enhance Troponin Immunoassay as Optimal Diagnostic Approach, Journal of Fish Diseases, 2025
Further publications are expected.
Additional information
- Researchers get to the heart of fish health with new diagnostic tool (fishfocus.co.uk)
- Diagnostic tool designed to improve fish heart health (fishfarmingexpert.com)
- Developing a mass testing tool for fish heart diseases | The Fish Site
- University of Glasgow - University news - Archive of news - 2021 - September - Researchers get to the heart of fish health with new diagnostic tool
- Diagnostic tool could help spot heart trouble in fish - Fish Farmer Magazine
- Heart of the matter - Fish Farmer Magazine
- Researchers get to the heart of fish health with new diagnostic tool (aquahoy.com)