Genomic breeding for gill health and lice resistance in salmon: improved accuracy and affordability
This project sought to optimise genomic selection in salmon breeding to improve the accuracy and affordability of the technology, with an end goal of applying these innovations to advance the breeding of salmon stocks with increased gill health and resistance to sea lice.
Project Summary
Project life: 48 months
Benefits
The project achieved its primary aim and objectives with significant developments in breeding for disease resistance, with a focus on gill health and sea lice traits. Cost-effective methods for genomic selection using low-density genetic markers were developed for sea lice resistance, and were extended to other target traits and species.
£1.07m
Total Value
56%
Industry Contribution
35%
SAIC Contribution
9%
Academia Contribution
Case study
Download full Case Study PDF
Partners
- Hendrix Genetics (formerly Landcatch Natural Selection)
- The Roslin Institute
- The Institute of Aquaculture at the University of Stirling
- Noahgene Ltd
Hendrix Genetics | University of Edinburgh | University of Stirling
Prof James Bron, Prof Ross Houston, Dr Smaragda Tsairidou, Dr Alastair Hamilton, Dr Halina Sobolewska, Dr Sophie Fridman and Dr Tharangani Herath.
Background
Cost-effective genomic selection has major potential to produce Atlantic salmon with increased disease resistance, thereby contributing to the continued sustainable development of the aquaculture sector. Sea lice mitigation costs the Scottish salmon sector approximately £27m per year, in terms of treatment costs and associated performance losses due to affected fish. Furthermore, amoebic gill disease (AGD), along with other gill diseases, pose a significant economic and welfare burden, particularly with the increasing water temperatures associated with climate change.
Methods for improving disease resistance are high priority areas for the global aquaculture industry with Scottish-based companies and universities at the forefront of developing these tools; however, wide-scale implementation remains prohibitively expensive for much of the sector.
Aims
The primary objective of this project was to optimise genomic selection in salmon breeding to improve the accuracy and affordability of the technology, with an end goal of applying these innovations to advance the breeding of salmon stocks with increased gill health and resistance to sea lice. The optimisation will include (1) the use of genotype imputation to make a step-reduction in cost of genomic selection, and (2) the inclusion of functional genomic variants within the genomic selection panels.
To achieve this, the following aims were established:
- To collect trait data related to resistance to lice (L. salmonis in Scotland) and (C. rogercresseyi) and AGD in large populations of pedigreed Atlantic salmon.
- To generate cost-efficient genome-wide genetic marker information for disease-challenged individuals and disease-free selection candidates.
- To test and optimize genomic prediction of breeding values for resistance to lice and AGD.
- To assess genetic correlations between resistance to the different parasites.
- To detect individual genes and mutations in the salmon genome affecting resistance to lice and AGD.
- To develop and apply genomic selection for multi-trait disease resistance in commercial breeding, and to benchmark the improvements versus conventional family methods.
- To actively disseminate results and technology to the aquaculture breeding and production sector, through publication and participation in trade and academic meetings and conferences.
The results of the project have been widely and rapidly disseminated across the sector to benefit aquaculture breeding and production.
Optimising methods of genomic selection for disease resistance traits in salmon
Based on Work Package (WP) 1: Collection of informative trait data and samples from disease challenge experiments; WP2: Generation of genome-wide marker data for trait-recorded fish; and WP3: Testing and optimising genomic selection for disease resistance.
Background
For successful implementation of genomic selection, there are two groups of individuals for which genome-wide marker information must be available:
- Individuals with the (disease) traits measured (aka training or reference population).
- Broodstock selection candidates which do not have disease measurements.
Genomic selection uses genomic and trait information on the former (i) to predict the disease resistance of the latter (ii) based on genomic information only. To test the effectiveness of genomic selection in the long term it is important to assess response to selection, but this is not practical within a single project. Therefore, in the short term it is possible to test the accuracy of breeding value predictions using genomic data by using large populations of individual fish with genome-wide genotype data and trait measurements. Analyses are then typically performed whereby subsets of the individuals have their trait measurements masked, and the accuracy of predicting those measurements using genotype data alone is assessed.
Work done
Analyses were performed using data and samples collected by Hendrix Genetics as part of their breeding programme operations and previous projects.
Sea lice challenge:
The dataset on which the majority of analyses were focused was a sea lice challenge, for which all samples were genotyped for genome-wide genetic markers using a Single-nucleotide Polymorphism (SNP) array. The primary aim of the study was to optimise the use of low-density genotypes and evaluate genotype imputation as a cost-effective method of genomic prediction. Imputation allows to predict high density genotypes from low density SNP panels, using information from haplotypes that are shared between related individuals. Phenotypes and genotypes (78,362 SNPs) were obtained for 610 individuals challenged with sea lice, Lepeophtheirus salmonis. The genomic prediction accuracy of genomic selection was calculated using genomic best linear unbiased prediction (gBLUP) approaches and compared across SNP panels of varying densities and composition, with and without imputation. Imputation was tested when parents were genotyped for the optimal SNP panel, and offspring were genotyped for a range of lower density imputation panels.
Reducing SNP density had little impact on prediction accuracy until 5,000 SNPs, below which the accuracy dropped. Imputation accuracy increased with increasing imputation panel density. Genomic prediction accuracy when offspring were genotyped for just 200 SNPs and imputed, and parents for 5,000 SNPs, was 0.53. This accuracy was similar to the full high density and optimal density dataset, and markedly higher than using 200 SNPs without imputation (Figure 1).
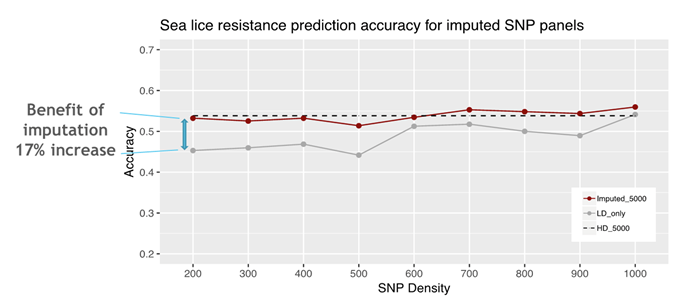
Figure 1: Prediction accuracy of imputation SNP panels for sea lice resistance for (i) the imputation SNP panel densities with range from 200 to 1,000 SNPs imputed to the optimal 5,000 SNP panel (Imputed_5000), (ii) imputation SNP panel densities with range from 200 to 1,000 SNPs alone without imputation (LD_only), and (iii) the optimal 5,000 SNP panel directly genotyped without imputation (HD_5000).
These results suggest that imputation from very low to medium density can be a cost-effective tool for genomic selection in Atlantic salmon breeding programs.
Unfortunately, the lack of availability of the required data and samples at the time of the project precluded the inclusion of the Chilean species Caligus rogercresseyi as originally planned. The original objective was to employ C. rogercresseyi data to provide additional understanding of the roots of sea louse resistance traits, but this could not be achieved. Caligus elongatus was not part of the original plan as large-scale infection challenges are difficult to achieve and on-farm infections in Scotland tend to be highly seasonal and to comprise mixed louse species, which reduces the ability to specifically identify C. elongatus impacts.
Gill health and other traits:
Similar methods were tested for gill health traits in salmon, and other target traits for other aquaculture species. The idea here was to test the potential for widespread adoption of these low-cost methods for broader uptake in aquaculture globally. The utility of low and medium density SNP panels (ranging from 100 to 9,000 SNPs) to accurately predict breeding values was tested and compared in four aquaculture datasets with different characteristics (species, genome size, genotyping platform, family number and size, total population size, and target trait).
The traits show heritabilities between 0.19-0.49, and genomic prediction accuracies using the full density panel of 0.55-0.87. A consistent pattern of genomic prediction accuracy was observed across species with little or no accuracy reduction until SNP density was reduced below 1,000 SNPs (prediction accuracies of 0.44-0.75). Below this SNP density, heritability estimates and genomic prediction accuracies tended to be lower and more variable (93% of maximum accuracy achieved with 1,000 SNPs, 89% with 500 SNPs, and 70% with 100 SNPs). A notable drop in accuracy was observed between 200 SNP panels (0.44-0.75) and 100 SNP panels (0.39-0.66) (Figure 2).
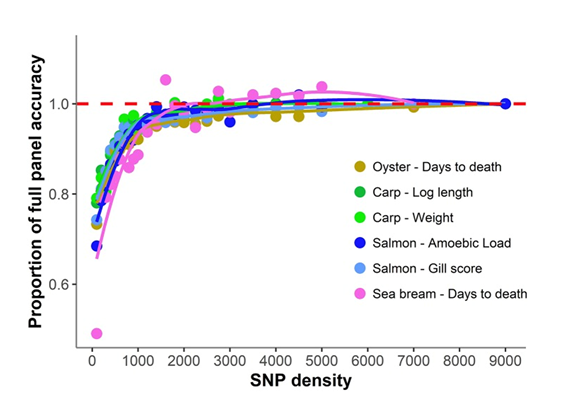
Figure 2: Proportion of genomic prediction accuracy achieved with low-density panels. The proportion of accuracy achieved by each SNP density was calculated by dividing the mean accuracy at that density by the mean accuracy obtained using the full high density SNP panels. The trend line was calculated using a Loess regression (local polynomial regression, span = 0.75).
Outcomes
Now that a multitude of studies have highlighted the benefits of genomic over pedigree-based prediction of breeding values in aquaculture species, the results of the current study highlight that these benefits can be achieved at lower SNP densities and at lower cost, raising the possibility of a broader application of genetic improvement in smaller and more fragmented aquaculture settings.
These results helped inform application of low-cost genomic selection in Hendrix Genetics breeding programmes and facilitated testing of genomic selection methods in other species, for example Pacific oysters, where imputation was demonstrated to be an effective tool for assisting with breeding value prediction at very low marker density. It is anticipated that further transfer of this technology to other aquaculture sectors will occur in coming years, as smaller and more fragmented industries are unlikely to be able to afford genotyping with high density SNP arrays.
Characterising disease resistance traits, genes, and potential causative variants in the salmon genome
Based on WP1: Collection of informative trait data and samples from disease challenge experiments; WP2: Generation of genome-wide marker data for trait-recorded fish; and WP4: Identifying disease resistance genes and causative variants in the salmon genome.
Background
While genetic variation in gill health and sea lice resistance traits has been clearly demonstrated, the mechanisms underlying genetic resistance are poorly characterised. Furthermore, breeding for disease resistance is typically performed using rather crude target traits such as gill health scores, or sea lice counts. Improved knowledge of the functional basis of genetic resistance can lead to identification of functional variants which could improve selection accuracy, but also help inform new target traits to include in the breeding goal.
Work done
One aim within this project was the characterisation of gene expression differences between farmed salmon of disparate genetic resistance based on amoebic gill disease challenges. RNA was extracted from the gill and head kidney of AGD resistant and susceptible animals following a challenge with N. perurans, and sequenced.
Outcomes
Comparison between resistant and susceptible animals primarily highlighted differences in the local immune response in the gill, involving red blood cell genes and genes related to immune function and cell adhesion. Differentially expressed immune genes pointed to a contrast in Th2 and Th17 responses, which is consistent with the increased heritability observed after successive challenges with the amoeba (Figure 3).
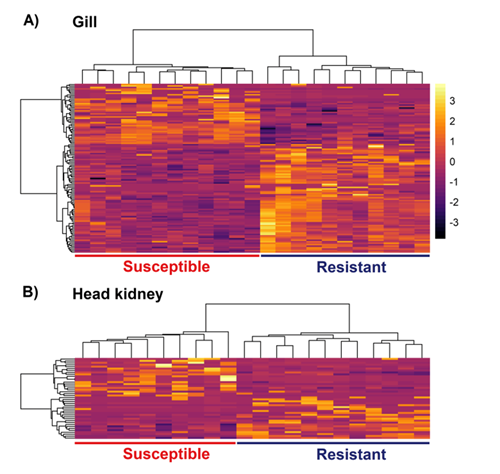
Figure 3: Heatmap of differentially expressed genes between resistant and susceptible samples. Heatmaps of all differentially expressed genes in gill (a) and head kidney (b). Samples and genes were clustered according to gene expression (mean centred and scaled normalised counts)
Five Quantitative trait locus (QTL) region candidate genes showed differential expression, including a gene connected to interferon responses (GVINP1), a gene involved in systemic inflammation (MAP4K4), and a positive regulator of apoptosis (TRIM39). Analyses of allele-specific expression highlighted a gene in the QTL region on chromosome 17, cellular repressor of E1A-stimulated genes 1 (CREG1), showing allelic differential expression suggestive of a cis-acting regulatory variant.
These results provide new insights into the mechanisms of resistance to AGD in Atlantic salmon, and highlights candidate genes for further functional studies that can further elucidate the genomic mechanisms leading to resistance and contribute to enhancing salmon health via improved genomic selection.
Development of novel, field-based, standardised gill scoring methodology for AGD
Based on WP1: Collection of informative trait data and samples from disease challenge experiments
Background
The final component of this work was characterisation of an emerging gill health issue, based on a cohort of farmed salmon which experienced gill health issues in Chile (from a company production site for which Hendrix Genetics provide genetic management). The aetiology of gill disorders is often considered to be multifactorial. Effective diagnosis, control and prevention are hindered by the lack of standardised methodologies to characterise the aetiological agents, which produce an array of clinical and pathological presentations. This also impacts on the use of gill scores as target traits for genetic improvement.
Work done
The aim of this study was to define a novel gross pathological scoring system suitable for field-based macroscopic assessment of complex or multifactorial gill disease in farmed Atlantic salmon, using samples derived from a gill disease outbreak in Chile. Clinical assessment of gross gill morphology was performed, and gill samples were collected for qPCR and histology.
Outcomes
A novel total gill scoring system was developed, which assesses gross pathological changes combining both the presumptive or healed AGD and the presence of other types of gill lesions (Table 1). This scoring system offers a standardised approach to characterise the severe proliferative pathologies in affected gills. Furthermore, this gill scoring system can substantially contribute to the development of robust mitigation strategies and could be used as an indicator trait for incorporating resistance to multifactorial gill disease into breeding goals.
Table 1. Total gill scoring system for gills of Atlantic salmon displaying multifactorial gill disease:
|
0 |
Clear healthy gill. |
|
1 |
Very light: discrete focal white streaks or patches on individual filaments and slight erosion/damage to distal ends of filaments. |
|
2 |
Light: more extensive coalescing white streaks or white focal patches on filaments, extended erosion/damage to distal ends of filaments. |
|
3 |
Moderate: extensive multi-filamental peripheral erosion, grossly swollen or thickened filaments (D: arrow) with occasional areas of localised areas of necrotic tissue. |
|
4 |
Advanced: extensive, grossly swollen or thickened filaments, shortened filaments (>50% of filament length affected) with areas of necrotic tissue. |
|
5 |
Severe: widespread necrotic patches, extensive melanisation, almost total destruction of gill architecture due to extensive loss of tissue. |
Impact
Based on WP6: Disseminating results and technology to the wider aquaculture sector.
The project achieved its primary aim and objectives with significant developments in breeding for disease resistance, with a focus on gill health and sea lice traits. Cost-effective methods for genomic selection using low density genetic markers were developed for sea lice resistance (https://pubmed.ncbi.nlm.nih.gov/28250015/), and was extended to other target traits and species (https://pubmed.ncbi.nlm.nih.gov/32174974/). The former was particularly notable for its development of genotype imputation methods to provide a very low-cost genotyping strategy for salmon breeding, and results received significant press attention (https://www.heraldscotland.com/news/18372164.super-salmon-project-end-scourge-sea-lice/). The results highlighted that similar accuracy of breeding value prediction could be achieved when the majority of animals were genotyped for just 200 genetic markers as when they were genotyped for tens of thousands of markers. In addition, the outputs of the project included detailed characterisation of amoebic gill disease host response in salmon using RNA sequencing methods, comparing resistant and susceptible animals (https://pubmed.ncbi.nlm.nih.gov/32228433/). Furthermore, a new gill disease syndrome was recently characterised as a case report (https://www.mdpi.com/2076-2607/9/12/2605), as a potential target trait for improvement of genetic resistance in the future.
The results of the project have been widely and rapidly disseminated across the sector to benefit aquaculture breeding and production, including via journal publications, conference presentations, and targeted press releases. The lead commercial partner, Hendrix Genetics, has used the results to inform their cutting-edge breeding programmes in Scotland and Chile and has been able to produce targeted genotyping panels for genomic selection at lower cost as a result of the project outputs. While selective breeding and genetic improvement have gradual and long-term benefit, the impacts of improved selection for sea lice and gill health traits are resulting in more robust salmon strains with higher levels of disease resistance. Finally, the consortium has also made significant contributions to important review papers on genetic technologies in UK aquaculture (e.g. https://onlinelibrary.wiley.com/doi/10.1111/raq.12553), and have used the SAIC project partnership to achieve UK and international funding to support additional research programmes targeting improvement of fish health and performance using genetic technologies.
Hendrix Genetics has used the results to inform its cutting-edge breeding programmes in Scotland and Chile.
The impacts of improved selection for sea lice and gill health traits are resulting in more robust salmon strains with higher levels of disease resistance.
Additional information
PRESENTATIONS
- Plant and Animal Genome Conference in January 2020 in San Diego, USA (by Ross Houston)
- Fourth Integrative Salmonid Biology Conference in Edinburgh in November 2019 (by Smaragda Tsairidou)
- Aquaculture Europe conference in Funchal in October 2021 (by Smaragda Tsairidou)
ARTICLES
Tsairidou, S., Hamilton, A., Robledo, D., Bron, J. E., & Houston, R. D., (2020). Optimizing low-cost genotyping and imputation strategies for genomic selection in Atlantic salmon. G3 (Bethesda), 10:581-590.
Kriaridou, C., Tsairidou, S., Houston, R. D., & Robledo, D., (2020). Genomic Prediction Using Low Density Marker Panels in Aquaculture: Performance Across Species, Traits, and Genotyping Platforms. Frontiers in genetics, 11, 124.
Robledo, D., Hamilton, A., Gutiérrez, A.P., Bron, J.E., & Houston, R.D., (2020). Characterising the mechanisms underlying genetic resistance to amoebic gill disease in Atlantic salmon using RNA sequencing. BMC Genomics 21(1):271.
Regan, T., Bean, T.P., Ellis, T., Davie, A., Carboni, S., Migaud, H., & Houston, R.D., (2021), Genetic improvement technologies to support the sustainable growth of UK aquaculture. Rev Aquacult, 13: 1958-1985.
Fridman, S., Tsairidou, S., Jayasuriya, N., Sobolewska, H., Hamilton, A., Lobos, C., Houston, R.D., Rodger, H., Bron, J. & Herath, T., (2021). Assessment of Marine Gill Disease in Farmed Atlantic Salmon (Salmo salar) in Chile Using a Novel Total Gross Gill Scoring System: A Case Study. Microorganisms. 9(12):2605.
PRESS RELEASES
- The Herald: ‘Super’ salmon project could end the scourge of sea lice
- Aquahoy: Low-cost method helps tackle sea lice in salmon
- The Fish Site: Cutting the cost of sea lice resistance in salmon
- Feedstuffs: Low-cost method helps tackle sea lice resistance in salmon
- Fish Farming Expert: Cut-price genetics used in search for sea lice resistant salmon
- Hatchery Feed and Management: Selective breeding to improve salmon resistance to sea lice
- Fish Focus: Aquaculture experts hatch new plan for sea lice resistance
The consortium has made significant contributions to important review papers on genetic technologies in UK aquaculture.
You will find an academic paper relating to this project, authored by Ross Houston and Dan Macqueen, here: Atlantic salmon (Salmo salar L.) genetics in the 21st century: taking leaps forward in aquaculture and biological understanding