Improving Disease Resistance to Flavobacterium psychrophilum
Improving disease resistance in Atlantic salmon and Rainbow trout to Flavobacterium psychrophilum, the causative agent of Rainbow Trout Fry Syndrome (RTFS).
Project Summary
Project life: 48 months
Benefits
This project, led by AquaGen Scotland, brought together research partners at the University of Stirling’s Institute of Aquaculture with producers Cooke Aquaculture and Dawnfresh to successfully develop and apply a new F. psychrophilum immersion challenge model to Atlantic salmon to enable assessment of susceptibility to disease and identification of DNA-markers for RTFS-resistance.
£827,264
Total Value
52%
Industry Contribution
38%
SAIC Contribution
10%
Academia Contribution
Partners
Partners
AquaGen Scotland Ltd, University of Stirling, Cooke Aquaculture Scotland, Dawnfresh Farming
Project Leads
Andrew Reeve, Fabian Grammes, Rowena Hoare, Valeria Macchia, Keng P Ang, Richard Hopewell
Case study
Download PDF
This project represented a good example of collaborative applied research, where the relationship between risk and reward suggested the need of public funding support. Here, public and private entities lead in their own field (AquaGen: breeding and genetics of salmonids; University of Stirling: aquaculture and molecular biology; Cooke Aquaculture Scotland and Dawnfresh Farming: salmon and trout farming, respectively) came together on a joint effort to solve a problem affecting the aquaculture sector in Scotland. … Together, and with the support from SAIC, these entities managed to bring more value to the industry; not just in Scotland but worldwide.
Project partners
Benefits
This project achieved its primary objectives by developing a new Flavobacterium psychrophilum immersion challenge model used to test both Atlantic salmon and Rainbow trout fry for their natural resistance to F. psychrophilum. Use of the immersion challenge model allowed for the successful identification of two strong QTL for resistance against F. psychrophilum in Atlantic salmon and contributed to one QTL in Rainbow trout to be used for marker assisted selection – selection of eggs that, because of the QTL genotypes of their parents, are expected to have inherited an increased resistance against F. psychrophilum.
BACKGROUND
Rainbow Trout Fry Syndrome (RTFS) has been responsible for substantial economic losses in the Rainbow trout sector globally, including the UK, for decades. RTFS, caused by the bacterium F. psychrophilum, occurs between 10-14°C, results in necrotic lesions on the skin surrounding the dorsal fin and tail and can cause high mortality in fry and larger fish in freshwater hatcheries and on-growing sites. Recently, F. psychrophilum has also been isolated from Atlantic salmon fry (<1g) in Scotland following disease outbreaks. Commercial vaccines against RTFS are currently unavailable in the UK, leaving antibiotics as the main treatment with increased reliance on a single antibiotic (florfenicol) to control the disease. Due to the potential risk of antibiotic resistance, the development of prophylactic measures to prevent RTFS, such as selective breeding for disease resistance and vaccine development are urgently required.
Understanding and exploiting the natural resistance of fish against disease is an effective mitigation approach for producing more robust populations. AquaGen, Lead Commercial Partner for this project, has previously identified and implemented genetic markers for disease resistance in Atlantic salmon to diseases including IPN, PD, CMS, HSMI and SRS. Furthermore, marker-assisted selection for F. psychrophilum resistance has recently been implemented in AquaGen’s production of Rainbow trout, indicating the potential for Atlantic salmon.
Until now, the lack of an effective and reproducible immersion challenge model capable of mimicking the natural route of infection has hampered the investigation of RTFS in Atlantic salmon. This project, led by AquaGen Scotland, brought together research partners at the University of Stirling’s Institute of Aquaculture with producers Cooke Aquaculture Scotland and Dawnfresh Farming to successfully develop and apply a new F. psychrophilum immersion challenge model to Atlantic salmon to enable assessment of susceptibility to disease and identification of DNA-markers for RTFS-resistance.
Outcomes from this project and ongoing work will result in the availability of eggs for the Atlantic salmon market in Scotland with increased disease resistance, as well as providing crucial information on susceptibility of RTFS resistant Rainbow trout to other disease. The industry value of this work is multifaceted, providing economic relief of the impacts of disease for the farmer while reducing the overall reliance on antibiotics as well as improving fish health and welfare throughout the production cycle.
AIMS
The primary aim of this project was to identify genetic markers associated with resistance against F. psychrophilum in Atlantic salmon. These markers were then used to select parent individuals for production of eggs with an inherited increased disease resistance.
Alongside this work, using the established immersion challenge model, research teams investigated the susceptibility of RTFS resistant Rainbow trout deployed on fish farms to other economically significant diseases, such as Proliferative Kidney Disease (PKD) and Red Mark Syndrome (RMS).
To achieve this, the following objectives were established:
1) To develop and apply an in vivo F. psychrophilum immersion challenge model in Atlantic salmon to enable assessment of susceptibility of families to disease.
2) To use material generated by immersion challenge trial to search for markers of resistance.
3) To produce salmon eggs selected for increased resistance to F. psychrophilum.
4) To perform a field trial to assess practical application of resistance selected stock on a Scottish fish farm and to genotype fish in the event of a F. psychrophilum disease outbreak.
5) To determine the susceptibility of RTFS resistant Rainbow trout deployed on fish farms to other economically significant diseases.
Development and application of in vivo disease challenge model
The identification of naturally resistant individuals requires the development and standardisation of an in vivo disease challenge model, specially designed for salmon, that closely mimics the natural exposure route of the pathogen. Project partners at the University of Stirling, having previously developed a successful challenge model for Rainbow trout, adapted and applied similar methodology for Atlantic salmon. Once this model was developed, families were challenged, and samples collected from resistant and susceptible fish to be used for genetic marker identification.
Development of the immersion challenge model
WORK DONE
Following previously developed Rainbow trout immersion model: pre-treatment with hydrogen peroxide was administered, followed by a seven-day post treatment observation period.
A safety test using unwashed bacterial isolates for immersion challenge was performed in two batches of Atlantic salmon fry.
The virulence of the F. psychrophilum isolates used is crucial, therefore fresh clinical isolates were provided by Cooke Aquaculture Scotland to be used for the following challenge experiment.
Three F. psychrophilum isolates were ‘passaged’ through fish to increase virulence by immersion and intramuscular injection in conjunction with a dose response experiment that determined the optimal dose for the disease susceptibility trial.
OUTCOMES
F. psychrophilum immersion challenge model was successfully developed and applied to Atlantic salmon fry. All fish survived the initial hydrogen peroxide pre-treatment and the immersion challenge with unwashed bacteria. Mortality resulting from the immersion challenge, confirmed to be caused by F. psychrophilum through PCR analysis, started three days post-challenge (dpc) and continued to 9 dpc with an average mortality rate of 36.6%.
Immersion challenge experiment
WORK DONE
Using the F. psychrophilum challenge model developed with the University of Stirling (UoS), an immersion challenge trial was performed in 2018 at VESO on Atlantic salmon fry from 200 families.
The trial was completed in duplicate tanks with fry of 1.1g and fish were pre-treated with hydrogen peroxide before challenged by immersion with F. psychrophilum.
Dead or moribund fish were collected, and fin clipped for DNA extraction. Specific mortality was verified by re-isolation of F. psychrophilum and PCR confirmed.
The trial was terminated after 23 days, and all remaining fish were assigned the status “survived”. All survivors were euthanised and DNA samples taken. The challenge was repeated in Spring of 2019, conducted in the same manner as the previous immersion challenge, and terminated after 20 days.
OUTCOMES
Initial (2018) and repeat (2019) immersion trials were successful at infecting Atlantic salmon fry with F. psychrophilum and both showed similar levels of infection and mortality trends between replicate tanks.
For the 2018 immersion trial the cumulative mortality at 23 days post infection (dpi) was 27.1% in tank 1 and 29.0% in tank 2 (Figure 1). Overall cumulative mortality was slightly lower for 2019 immersion trial (18.1%).
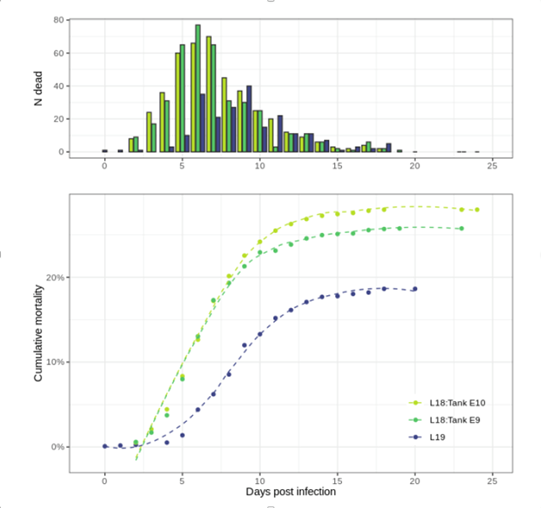
Figure 1: F. psychrophilum induced mortalities in the Atlantic salmon challenges in 2018 (L18) and 2019 (L19): The upper panel shows the number of dead fish by days post-infection. The lower panel shows the cumulative mortality in percent by days post infection.
Identification of QTL & marker-assisted selection of resistant fish
The main objective of this work package was to determine the degree of genetic variation in disease resistance in Atlantic salmon and to identify quantitative trait loci (QTL) associated with resistance to infection by F. psychrophilum. A QTL is a region of DNA associated with a particular phenotypic trait which can be used for marker-assisted selection (MAS).
Identification of QTL in Atlantic salmon
WORK DONE
DNA extractions were performed by Vaxxinova Norway using fin clip samples from the 2018/2019 immersion challenge trials and all individuals genotyped using AquaGen’s 70k SNP-chip based on Affymetrix technology.
Raw genotyping data of the samples from both year classes were processed independently as batches which resulted in filtered datasets containing 53809 SNPs and 57450 SNPs for 2018 and 2019, respectively. All SNPs were placed on the Atlantic salmon genome ICSASGv2 (GCF_000233375.1).
OUTCOMES
Analysis revealed a significant genetic component for resistance against F. psychrophilum in Atlantic salmon through estimates of genetic heritability (h2), i.e., the proportion of phenotypic variance explained by the SNPs, for 2018 (h2 = 0.381) and 2019 (h2 = 0.173). Genetic correlation (rG) for the trait survival was high between both tanks used in the challenge for 2018 (rG=0.959 ± 0.041). Similarly, a high genetic correlation (rG=0.989 ± 0.081) was found for survival between the challenges from both year classes, indicating general, high, reproducibility of the results.
A further genome wide association study (GWAS) was conducted separately on the 2018/19 datasets (Figure 2). The results were consistent and clearly indicated two strong QTLs located on chromosome 5 (QTL5) and 9 (QTL9) with the vast majority of significant SNPs being located on these two chromosomes. The top SNPs from each of the chromosomes were identical between 2018/2019 challenge trials and consistent across year classes; thus, confirming the existence of two QTLs. Both QTLs showed epistasis, i.e., interaction between genes, with an increased chance of survival for individuals possessing the “good” alleles from both QTLs (Figure 3). This translates to an estimated 90% increase in relative percent survival by selecting for both QTLs.
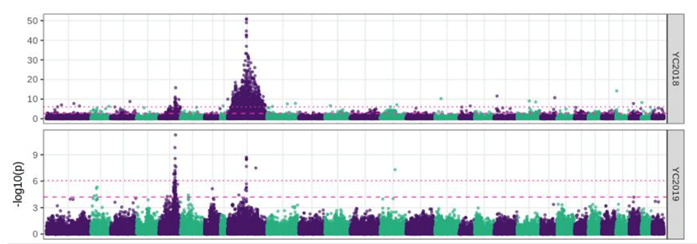
Figure 2: Manhattan plots for genome-wide association analysis of F. psychrophilum resistance of the 2018 (top) and 2019 (bottom). The figure shows the -log10 (p-value) of the test statistics for each SNP plotted against the physical positions of the markers on the chromosomes. The two lines in each panel indicate significance threshold after accounting for multiple-testing using either: Bonferroni correction (dotted line) or False Discovery Rate correction (dashed line).
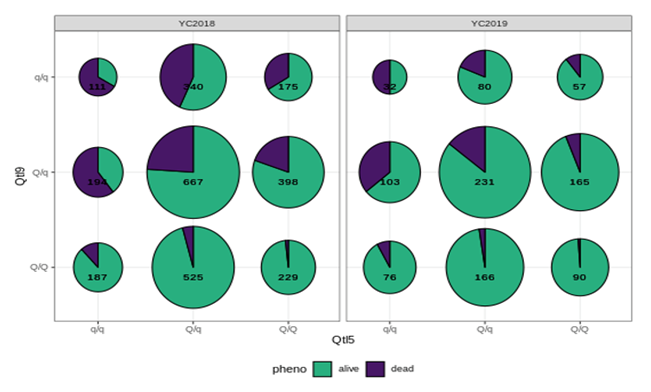
Figure 3: Epistatic effects between the 2 QTLs located on chromosome 5 and 9 in the two year classes (YC)2018 and 19, shown as bubble plots. The genotypes for Qtl5 and 9 are plotted on the x- and y-axis, the size of the bubbles is proportional to the number of animals with that combination of genotypes (displayed as number in the bubble). “Q/q” represent a copy of the good/bad allele of the respective QTL.
Field trial and commercial implementation
The next logical step was to validate the effect of the newly identified QTLs for RTFS resistance in a commercial setting by genotyping both resistant (survivors) and susceptible (mortalities) individuals following a natural outbreak F. psychrophilum. Due to unavoidable circumstances, field validation was not possible within the project timeline and remains to be completed. Despite this, resistance to F. psychrophilum has been implemented by the project leader AquaGen as part of its add-on products, and batches of eggs with this characteristic can be produced when customers request them.
Additional benefit: Project outputs derive added value for Scottish Rainbow trout industry
Alongside ongoing work, a series of trials were initiated in Rainbow trout to maximise output from material generated by the project.
Immersion challenge and identification of QTL in trout
The newly developed F. psychrophilum immersion challenge model was trialled on Rainbow trout at VESO Vikan, Norway. Two challenges were conducted; the first in 2019 using 2000 (2g) trout fry supplied by AquaGen and the second in 2020 using 3000 fry (1.1g) supplied as a mixture from two sources AquaGen and AquaSearch. Comparable challenge methodologies and sampling regimes were used, and results indicated that the overall trend in cumulative mortality was similar to that in the salmon trials. At the end of the trials, resistant (surviving) and susceptible (mortalities) fish were sampled for DNA extraction and genotyping analysis carried out.
The estimates for genetic heritability (h2) for resistance against F. psychrophilum coincide with results from earlier challenge trials; 2019 (h2 = 0.37) and for the 2020 (h2= 0.32 for AquaGen population) and (h2 = 0.38 for AquaSearch population). Genetic correlation between the 2019 and the 2020 (AquaGen) challenge was 0.59+/-0.16, again, indicating good reproducibility of the challenge.
A subsequent GWAS was conducted separately on the 2019 challenge as well as each population of the 2020 challenge which led to the identification of a new QTL located on chromosome 25 for resistance against F. psychrophilum in Rainbow trout.
Monitoring infections in QTL Rainbow trout stock
Another section of work was undertaken to determine the susceptibility of RTFS resistant Rainbow trout deployed on fish farms to other economically significant diseases such as Proliferative Kidney Disease (PKD) and Red Mark Syndrome (RMS) using previously developed tests. The work was carried out as part of two MSc projects at the Institute of Aquaculture, University of Stirling, supervised by Prof. Sandra Adams and Dr Rowena Hoare.
Rainbow trout eggs provided by AquaGen were deployed on a DawnFresh farm site in 2017 and followed in a longitudinal study for the presence of F. psychrophilum, RMS and PKD and response to vaccination against Aeromonas salmonicida.
Fish were sampled over a three-month period for analysis by PCR, immunohistochemistry and ELISA. Following analysis there was no detection of RMS or PKD in either stock (RTFS resistant selected or commercial). However, 6% of selected fish and 7.2% of non-selected fish were positive for F. psychrophilum (confirmed by PCR) following an outbreak of F. psychrophilum that required antibiotic treatment.
No significant differences were found in antibody response to vaccination between the stocks. The use of immunohistochemistry to monitor gill chloride cells was successful. No difference was found in numbers of chloride cells between stocks. A rapid method to identify F. psychrophilum was developed: Immuno-fluorescent antibody technique (IFAT). Unfortunately, the fieldwork was terminated early owing to a commercial decision to transfer them to sea ahead of plan in response to an unforeseen issue.
In the case of the field trial for Rainbow trout, the data were unable to support differentiated stocks’ expected performance, and statistical analysis of the results showed no difference in terms of infection related mortality. This was a key finding for the industry partner, and an area for further consideration.
IMPACT
This project was successful in fully achieving its two main goals:
- Development and successful application of an in vivo psychrophilum challenge model for Atlantic salmon.
- Identification of two strong QTL for resistance against psychrophilum in Atlantic salmon.
The immersion challenge model developed during the project can be applied in future trials to examine the genetic component behind resistance to this disease in both salmon and trout. Furthermore, the challenge model has the potential to be utilised for the development of mucosal vaccines; both strategies of which will improve the sustainability of the industry and lead to an overall reduction in antibiotic usage.
The QTLs identified for F. psychrophilum resistance in salmon are already being used by AquaGen for marker assisted selection in the production RTFS resistant egg batches. Increasing disease resistance and overall fish health and welfare through selective breeding and stock improvement is a continual and evolving process. The ability to select for RTFS resistance is an important addition to the growing list of commercially important traits that can be incorporated into breeding programmes alongside other traits such as: Increased growth, delayed sexual maturation, and improved fillet quality, as well as resistance to a range of other diseases. Selection of these traits can be weighted, resulting in the ability to produce egg batches tailored for success in specific locations or conditions.
This project generated further learnings through supporting three academic projects (2 MSc projects and a PhD, co-funded by Stirling University and AquaGen) looking at both Atlantic salmon and Rainbow trout, resulting in a new rapid detection method for F. psychrophilum using IFAT and ELISA and a specific, sensitive, and cost-effective SYBER Green assay to detect F. psychrophilum in fish tissues. Improved methods of detection of F. psychrophilum (qPCR, IFAT) will aid in diagnosis and control of the disease. Furthermore, the results from this project contributed to the identification of a new QTL located on chromosome 25 for resistance against F. psychrophilum in Rainbow trout, allowing AquaGen to improve its selection of Rainbow trout eggs, too.
Although the initial scope of this work changed over the course of the four-year project, with field trials and validation not fully realised, the valued outcomes and impact to the sector are highly relevant and have been readily implemented.
The project represented a good example of collaborative applied research, where the relationship between risk and reward suggested the need of public funding support. Here, public and private entities lead in their own field (AquaGen: breeding and genetics of salmonids; University of Stirling: aquaculture and molecular biology; Cooke Aquaculture Scotland and Dawnfresh Farming: salmon and trout farming, respectively) came together on a joint effort to solve a problem affecting the aquaculture sector in Scotland. The positive and efficient collaboration allowed good initial discussions that where then translated into a productive planning and execution of activities. Together, and with the support from SAIC, these entities managed to bring more value to the industry; not just in Scotland but worldwide.
Additional information
Presentations:
Valeria Macchia, et al; IMPROVING DISEASE RESISTANCE IN RAINBOW TROUT TO FLAVOBACTERIUM PSYCHROPHILUM, THE CAUSATIVE AGENT OF RAINBOW TROUT FRY SYNDROME; 2019 BTA Annual Conference (September 18-19, 2019).
Fabian Grammes, et al; A MAJOR QTL FOR Flavobacterium psychrophilum RESISTANCE IN ATLANTIC SALMON (SALMO SALAR); Aquaculture Europe 2020 (Online, April 12-15, 2021).
Valeria Macchia, et al; QUANTIFICATION OF FLAVOBACTERIUM PSYCHROPHILUM IN IMMERSION CHALLENGED ATLANTIC SALMON (SALMO SALAR) SELECTED FOR DISEASE RESISTANCE; Aquaculture Europe 2020 (Online, April 12-15, 2021).
Valeria Macchia, et al; IMMUNOHISTOCHEMICAL DETECTION OF IGT-POSITIVE CELLS AND FLAVOBACTERIUM PSYCHROPHILUM IN IMMERSION CHALLENGED ATLANTIC SALMON (SALMO SALAR) (Online, September 20-23, 2021).
Fabian Grammes, et al; A MAJOR QTL FOR Flavobacterium psychrophilum RESISTANCE IN ATLANTIC SALMON (SALMO SALAR); LATAM (Latin America Quick Talks in Aquaculture Genomics) 2021.
Fabian Grammes, et al; A MAJOR QTL FOR Flavobacterium psychrophilum RESISTANCE IN ATLANTIC SALMON (SALMO SALAR); Aquaculture UK (Aviemore, Scotland 3-5 May 2022).
Publications:
Macchia, M. Inami, A. Ramstad, F. Grammes, A. Reeve, T. Moen, J.S. Torgersen, A. Adams, A.P. Desbois, R. Hoare: Immersion challenge model for Flavobacterium psychrophilum infection of Atlantic salmon (Salmo salar L.) fry. (Journal of Fish Diseases, 45(11), 1781-1788. https://doi.org/10.1111/jfd.13699)
Grammes, V. Macchia, A. Reeve, TM Knutson, T. Moen, A. Adams, F. Grammes, A. Desbois, R. Hoare: Two strong QTLs controlling resistance against Flavobacterium psychrophilum in Salmo salar. (Manuscript in preparation)
Mathiessen, Y. Duan, M.H. Marana, S. Zuo, A.M. Karami, R. Jafaar, L. von Gersdorff Jørgensen, P. W. Kania, I. Dalsgaard, L. Madsen, T. Nielsen, F. Grammes, J. Ødegård, V Macchia, K. Buchmann: Validation of a QTL for Flavobacterium psychrophilum resistance in rainbow trout Oncorhynchus mykiss (Aquaculture Reports, 2023, https://doi.org/10.1016/j.aqrep.2023.101573)
Press release:
“Improving disease resistance to Flavobacterium psychrophilum” (May 2021). Available on the following websites:
- https://www.fishfarmingexpert.com/article/genetic-discovery-points-the-way-to-flavobacteriosis-resistant-salmon/
- https://thefishsite.com/articles/genetic-breakthrough-could-save-farmed-salmon-from-flavobacteriosis
- https://www.fishfarmermagazine.com/news/aquagen-led-team-in-salmon-health-breakthrough/
- https://www.hatcheryfm.com/hfm-article/1418/Genetic-discovery-marks-significant-progress-in-fight-against-flavobacteriosis/
- https://salmonbusiness.com/significant-breakthrough-with-genetic-discovery-in-fight-against-fish-disease/
- https://www.undercurrentnews.com/2021/05/26/new-genetics-finding-could-be-lifeline-for-juvenile-salmon/
- https://fishfocus.co.uk/genetic-discovery-in-fight-against-fish-disease/